-Search query
-Search result
Showing all 45 items for (author: pramanick & i)
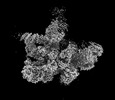
EMDB-18920: 
Plastid-encoded RNA polymerase (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
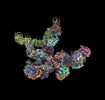
EMDB-18935: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18952: 
Plastid-encoded RNA polymerase transcription elongation complex (consensus map)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18964: 
Plastid-encoded RNA polymerase (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18965: 
Plastid-encoded RNA polymerase (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18974: 
Plastid-encoded RNA polymerase (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18975: 
Plastid-encoded RNA polymerase (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18976: 
Plastid-encoded RNA polymerase (Region 5)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18982: 
Plastid-encoded RNA polymerase (Region 6)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18983: 
Plastid-encoded RNA polymerase (Region 8)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18985: 
Plastid-encoded RNA polymerase (Region 9)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18986: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 1)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18995: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 2)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18996: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 3)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-18998: 
Plastid-encoded RNA polymerase (Region 7)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-19007: 
Plastid-encoded RNA polymerase transcription elongation complex (Region 4)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
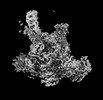
EMDB-19010: 
Plastid-encoded RNA polymerase transcription elongation complex (PAP2-mRNA)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r5o: 
Plastid-encoded RNA polymerase
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8r6s: 
Plastid-encoded RNA polymerase (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8rdj: 
Plastid-encoded RNA polymerase transcription elongation complex (Integrated model)
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A
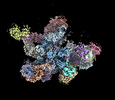
EMDB-19023: 
Plastid-encoded RNA polymerase transcription elongation complex
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

PDB-8ras: 
Plastid-encoded RNA polymerase transcription elongation complex
Method: single particle / : Webster MW, Pramanick I, Vergara-Cruces A

EMDB-33411: 
Supramolecular assembly of Thermostable Direct Hemolysin from Vibrio parahaemolyticus
Method: single particle / : Mishra S, Dutta S

EMDB-33412: 
Wild type Thermostable Direct Hemolysin (TDH) from Vibrio parahaemolyticus
Method: single particle / : Mishra S, Dutta S

EMDB-33413: 
N-terminal deleted (NTD) Thermostable Direct Hemolysin from Vibrio parahaemolyticus
Method: single particle / : Mishra S, Dutta S

EMDB-33331: 
Cyo-EM model for native cystathionine beta-synthase of Mycobacterium tuberculosis.
Method: single particle / : Bandyopadhyay P, Pramanick I, Biswas R, Sabarinath PS, Sreedharan S, Singh S, Rajmani R, Laxman S, Dutta S, Singh A
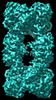
EMDB-33348: 
Cryo-EM map of cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine.
Method: single particle / : Bandyopadhyay P, Pramanick I, Biswas R, Sabarinath PS, Sreedharan S, Singh S, Rajmani R, Laxman S, Dutta S, Singh A
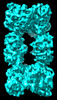
EMDB-33363: 
Cryo-EM map of cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine and serine.
Method: single particle / : Bandyopadhyay P, Pramanick I, Biswas R, Sabarinath PS, Sreedharan S, Singh S, Rajmani R, Laxman S, Dutta S, Singh A

PDB-7xnz: 
Native cystathionine beta-synthase of Mycobacterium tuberculosis.
Method: single particle / : Bandyopadhyay P, Pramanick I, Biswas R, Sabarinath PS, Sreedharan S, Singh S, Rajmani R, Laxman S, Dutta S, Singh A

PDB-7xoh: 
Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine.
Method: single particle / : Bandyopadhyay P, Pramanick I, Biswas R, Sabarinath PS, Sreedharan S, Singh S, Rajmani R, Laxman S, Dutta S, Singh A

PDB-7xoy: 
Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine and serine.
Method: single particle / : Bandyopadhyay P, Pramanick I, Biswas R, Sabarinath PS, Sreedharan S, Singh S, Rajmani R, Laxman S, Dutta S, Singh A

EMDB-32388: 
Cryo-EM 3D model of the 3-RBD up dimeric spike protein of SARS-CoV2 in the presence of SIH-5
Method: single particle / : Khatri B, Pramanick I, Malladi SK, Rajmani RS, Kumar S, Ghosh P, Sengupta N, Rahisuddin R, Kumaran S, Ringe RP, Varadarajan R, Dutta S, Chatterjee J
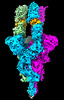
EMDB-33042: 
Cryo-EM 3D model of the 3-RBD up single trimeric spike protein of SARS-CoV2 in the presence of synthetic peptide SIH-5.
Method: single particle / : Khatri B, Pramanick I, Malladi SK, Rajmani RS, Kumar S, Ghosh P, Sengupta N, Rahisuddin R, Kumaran S, Ringe RP, Varadarajan R, Dutta S, Chatterjee J

PDB-7x7n: 
3D model of the 3-RBD up single trimeric spike protein of SARS-CoV2 in the presence of synthetic peptide SIH-5.
Method: single particle / : Khatri B, Pramanick I, Malladi SK, Rajmani RS, Kumar S, Ghosh P, Sengupta N, Rahisuddin R, Kumaran S, Ringe RP, Varadarajan R, Dutta S, Chatterjee J

EMDB-31092: 
SARS-CoV2 Spike Protein structure at pH 6.5 with C1 Symmetry (Class 2)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31093: 
SARS-CoV2 Spike Protein structure at pH 6.5 with C1 Symmetry (Class 3)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31094: 
SARS-CoV2 Spike Protein structure at pH 6.5 with C1 Symmetry (Class 4)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31095: 
SARS-CoV2 Spike Protein structure at pH 6.5 with C1 Symmetry (Class 5)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31096: 
SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 3)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31097: 
SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 5)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31098: 
SARS-CoV2 Spike Protein structure at pH 7.4 with C3 Symmetry
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31099: 
SARS-CoV2 Spike Protein structure at pH 8.0 with C1 Symmetry (Class 1)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31100: 
SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 9)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31101: 
SARS-CoV2 Spike Protein structure at pH 7.4 with C1 Symmetry (Class 8)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S

EMDB-31102: 
SARS-CoV2 Spike Protein structure at pH 8.0 with C1 Symmetry (Class 2)
Method: single particle / : Pramanick I, Sengupta N, Mishra S, Pandey S, Girish N, Das A, Dutta S
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
